-Search query
-Search result
Showing all 50 items for (author: turonova & b)
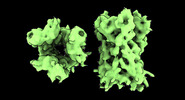
EMDB-17704: 
Subtomogram average of Vaccinia A10 trimer with open center from in vitro cores
Method: subtomogram averaging / : Turonova B, Liu J
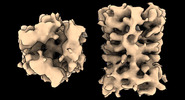
EMDB-17708: 
Subtomogram average of Vaccinia A10 trimer with tight center from in vitro cores
Method: subtomogram averaging / : Turonova B, Liu J

EMDB-17753: 
Subtomogram average of Vaccinia A10 trimer from in situ cores
Method: subtomogram averaging / : Turonova B, Liu J

EMDB-16721: 
Human 80S ribosome in untreated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16722: 
Human 80S ribosome in HHT-treated cells
Method: subtomogram averaging / : Xing H, Taniguchi R, Khusainov I, Kreysing JP, Welsch S, Turonova B, Beck M

EMDB-16725: 
Human 80S ribosome at the 'eEF1A, A/T, P' state in cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16726: 
Human 80S ribosome at the 'A/T, P' state in untreated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16727: 
Human 80S ribosome at the 'eEF2, ap/P, pe/E' state in untreated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16728: 
Human 80S ribosome at the 'eEF1A, A/T, P, E' state in untreated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16733: 
Human 80S ribosome at the 'eEF2' state in untreated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16734: 
Human 80S ribosome at the 'P' state in untreated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16735: 
Human 80S ribosome at the 'A, P' state in untreated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16736: 
Human 80S ribosome at the 'eEF1A, A/T, P, Z' state in untreated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16737: 
Human di-ribosome in untreated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16738: 
Human 80S ribosome with ES27L at conformation 1 in untreated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16739: 
Human 80S ribosome with ES27L at conformation 2 in untreated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16740: 
Human 80S ribosome with ES27L at conformation 3 in untreated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16741: 
Human 80S ribosome with ES27L at conformation 4 in untreated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16742: 
Human 80S ribosome with ES27L at conformation 5 in untreated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16743: 
Human 60S in the cytoplasm of untreated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16744: 
Human 80S ribosome at the 'eEF2' state in HHT-treated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16747: 
Human 80S ribosome at the 'A, P' state in HHT-treated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16748: 
Human 80S ribosome at the 'P' state in HHT-treated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16749: 
Human 80S ribosome at the 'eEF1A, A/T, P, Z' state in HHT-treated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16750: 
Human 80S ribosome at the 'A/T, P, Z' state in HHT-treated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16751: 
Human 80S ribosome at the potential 'Compact eEF2, A, P' state in HHT-treated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16752: 
Human 80S ribosome with ES27L at conformation 1 in HHT-treated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16754: 
Human 80S ribosome with ES27L at conformation 5 in HHT-treated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16755: 
Human 80S ribosome with ES27L at conformation 6 in HHT-treated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16756: 
Human 80S ribosome with ES27L at conformation 7 in HHT-treated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-16757: 
Human 60S associated with eIF6 in the cytoplasm of HHT-treated cells
Method: subtomogram averaging / : Xing H, Welsch S, Turonova B, Beck M

EMDB-14325: 
Cytoplasmic ring of the human nuclear pore complex from isolated HeLa nuclear envelopes
Method: subtomogram averaging / : Mosalaganti S, Obarska-Kosinska A, Siggel M, Taniguchi R, Turonova B, Zimmerli CE, Buczak K, Schmidt FH, Margiotta E, Mackmull MT, Hagen WJH, Hummer G, Kosinski J, Beck M

EMDB-14326: 
Structure of nuclear pore complex from HEK cells with GP210 knockout
Method: subtomogram averaging / : Mosalaganti S, Obarska-Kosinska A, Siggel M, Taniguchi R, Turonova B, Zimmerli CE, Buczak K, Schmidt FH, Margiotta E, Mackmull MT, Hagen WJH, Hummer G, Kosinski J, Beck M

EMDB-14327: 
Nuclear pore complex from intact HeLa cells
Method: subtomogram averaging / : Mosalaganti S, Obarska-Kosinska A, Siggel M, Taniguchi R, Turonova B, Zimmerli CE, Buczak K, Schmidt FH, Margiotta E, Mackmull MT, Hagen WJH, Hummer G, Kosinski J, Beck M

EMDB-14328: 
Inner/spoke ring of the human nuclear pore complex from isolated HeLa nuclear envelopes.
Method: subtomogram averaging / : Mosalaganti S, Obarska-Kosinska A, Siggel M, Taniguchi R, Turonova B, Zimmerli CE, Buczak K, Schmidt FH, Margiotta E, Mackmull MT, Hagen WJH, Hummer G, Kosinski J, Beck M

EMDB-14330: 
Nuclear ring of the human nuclear pore complex from isolated HeLa nuclear envelopes
Method: subtomogram averaging / : Mosalaganti S, Obarska-Kosinska A, Siggel M, Taniguchi R, Turonova B, Zimmerli CE, Buczak K, Schmidt FH, Margiotta E, Mackmull MT, Hagen WJH, Hummer G, Kosinski J, Beck M

EMDB-14321: 
Human nuclear pore complex (dilated)
Method: subtomogram averaging / : Mosalaganti S, Obarska-Kosinska A, Siggel M, Taniguchi R, Turonova B, Zimmerli CE, Buczak K, Schmidt FH, Margiotta E, Mackmull MT, Hagen WJH, Hummer G, Kosinski J, Beck M

PDB-7r5j: 
Human nuclear pore complex (dilated)
Method: subtomogram averaging / : Mosalaganti S, Obarska-Kosinska A, Siggel M, Taniguchi R, Turonova B, Zimmerli CE, Buczak K, Schmidt FH, Margiotta E, Mackmull MT, Hagen WJH, Hummer G, Kosinski J, Beck M

EMDB-14322: 
Human nuclear pore complex (constricted)
Method: subtomogram averaging / : Mosalaganti S, Obarska-Kosinska A, Siggel M, Taniguchi R, Turonova B, Zimmerli CE, Buczak K, Schmidt FH, Margiotta E, Mackmull MT, Hagen WJH, Hummer G, Kosinski J, Beck M

PDB-7r5k: 
Human nuclear pore complex (constricted)
Method: subtomogram averaging / : Mosalaganti S, Obarska-Kosinska A, Siggel M, Taniguchi R, Turonova B, Zimmerli CE, Buczak K, Schmidt FH, Margiotta E, Mackmull MT, Hagen WJH, Hummer G, Kosinski J, Beck M

EMDB-14243: 
cryoEM structure of human Nup155 (residues 19-981)
Method: single particle / : Taniguchi R, Beck M

EMDB-11222: 
Structure of SARS-CoV-2 spike glycoprotein (S) trimer determined by sub-tomogram averaging
Method: subtomogram averaging / : Turonova B, Sikora M, Schurmann C, Hagen W, Welsch S, Blanc FEC, von Bulow S, Gecht M, Bagola K, Horner C, van Zandbergen G, Landry J, de Azevedo NTD, Mosalaganti S, Schwarz A, Covino R, Muhlebach M, Hummer G, Locker JK, Beck M

EMDB-11223: 
Structure of SARS-CoV-2 spike glycoprotein (S) monomer in a closed conformation determined by sub-tomogram averaging
Method: subtomogram averaging / : Turonova B, Sikora M, Schurmann C, Hagen WJH, Welsch S, Blanc FEC, von Bulow S, Gecht M, Bagola K, Horner C, van Zandbergen G, Landry J, de Azevedo NTD, Mosalaganti S, Schwarz A, Covino R, Muhlebach M, Hummer G, Locker JK, Beck M

EMDB-11967: 
Human nuclear pore complex in HIV-1 infected T cells
Method: subtomogram averaging / : Margiotta E, Beck M

EMDB-11347: 
Structure of SARS-CoV-2 spike glycoprotein (S) trimer with one receptor binding domain (RBD) in open-state determined by subtomogram averaging
Method: subtomogram averaging / : Turonova B, Sikora M, Schurmann C, Hagen W, Welsch S, Blanc FEC, von Bulow S, Gecht M, Bagola K, Horner C, van Zandbergen G, Laundry J, de Azevedo NTD, Mosalaganti S, Schwarz A, Covino R, Muhlebach M, Hummer G, Locker JK, Beck M

EMDB-10660: 
Nup116_delta_NPC_25C
Method: subtomogram averaging / : Allegretti M, Zimmerli CE, Rantos V, Wilfling F, Ronchi P, Fung HKH, Lee CW, Hagen W, Turonova B, Karius K, Zhang X, Mulller C, Schwab Y, Mahamid J, Pfander B, Kosinski J, Beck M

EMDB-10661: 
Nup116delta_NPC_37C
Method: subtomogram averaging / : Allegretti M, Zimmerli CE, Rantos V, Wilfling F, Ronchi P, Fung HKH, Lee CW, Hagen W, Turonova B, Karius K, Zhang X, Muller C, Schwab Y, Mahamid J, Pfander B, Kosinski J, Beck M

EMDB-10198: 
In-cell S cerevisiae nuclear pore complex
Method: subtomogram averaging / : Beck M, Allegretti M

EMDB-10207: 
A 4.2 Angstrom structure of HIV-1 CA-SP1
Method: subtomogram averaging / : Turonova B, Hagen WJH, Obr M, Mosalaganti S, Kraeusslich HG, Beck M

EMDB-3782: 
A 3.4 Angstrom structure of HIV-1 CA-SP1 by 3D-CTF correction of cryo-electron tomograms
Method: subtomogram averaging / : Turonova B, Schur FKM, Wan W, Briggs JAG
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
